Note
Click here to download the full example code
Learning curves (layer widths)¶
Hello world
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense
from tensorflow.keras.initializers import GlorotUniform
from sklearn.datasets import load_boston
from sklearn.model_selection import train_test_split
from matplotlib.colors import LogNorm
from mpl_toolkits.mplot3d import Axes3D
from etudes.metrics import nmse
num_layers = 2
num_epochs = 100
batch_size = 64
val_rate = 0.2
seed = 8888 # set random seed for reproducibility
random_state = np.random.RandomState(seed)
t_grid = np.arange(num_epochs)
width_grid = np.arange(2, 65)
dataset = load_boston()
X_train, X_val, y_train, y_val = train_test_split(dataset.data, dataset.target,
test_size=val_rate,
random_state=random_state)
frames = []
for i, width in enumerate(width_grid):
num_units = int(width)
model = Sequential()
for _ in range(num_layers):
model.add(Dense(num_units, activation="relu",
kernel_initializer=GlorotUniform(seed=seed)))
model.add(Dense(1, kernel_initializer=GlorotUniform(seed=seed)))
model.compile(optimizer="adam", loss="mean_squared_error", metrics=[nmse])
hist = model.fit(X_train, y_train, validation_data=(X_val, y_val),
epochs=num_epochs, batch_size=batch_size, verbose=False)
frame = pd.DataFrame(hist.history).assign(width=width, seed=seed)
frame.index.name = "epoch"
frame.reset_index(inplace=True)
frames.append(frame)
data = pd.concat(frames, axis="index", ignore_index=True, sort=True)
data.rename(lambda s: s.replace('_', ' '), axis="columns", inplace=True)
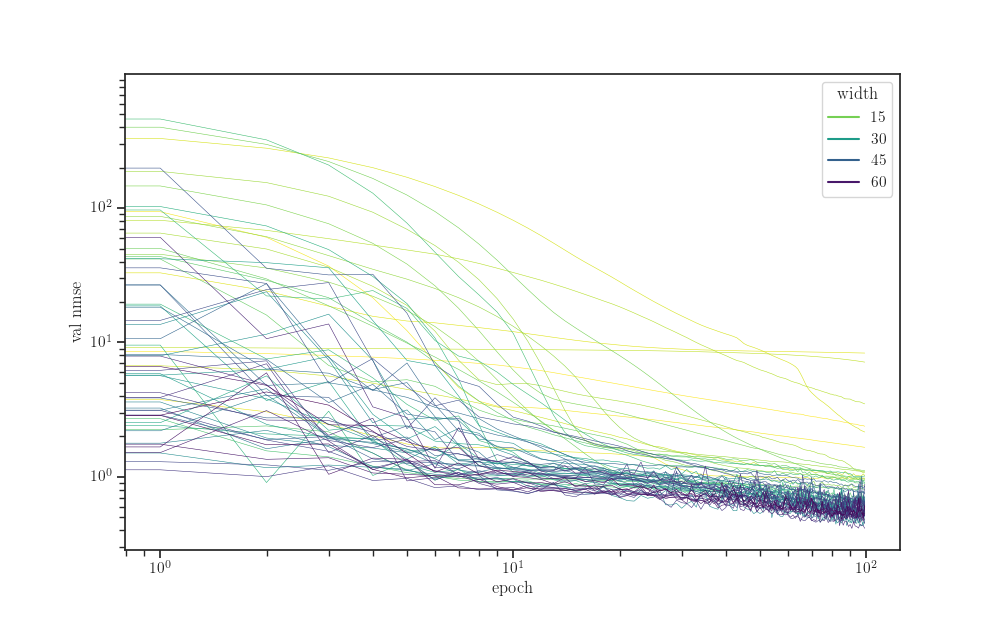
fig, ax = plt.subplots()
sns.lineplot(x="epoch", y="val nmse", hue="width",
units="seed", estimator=None,
palette="viridis_r", linewidth=0.4,
data=data, ax=ax)
ax.set_xscale("log")
ax.set_yscale("log")
plt.show()
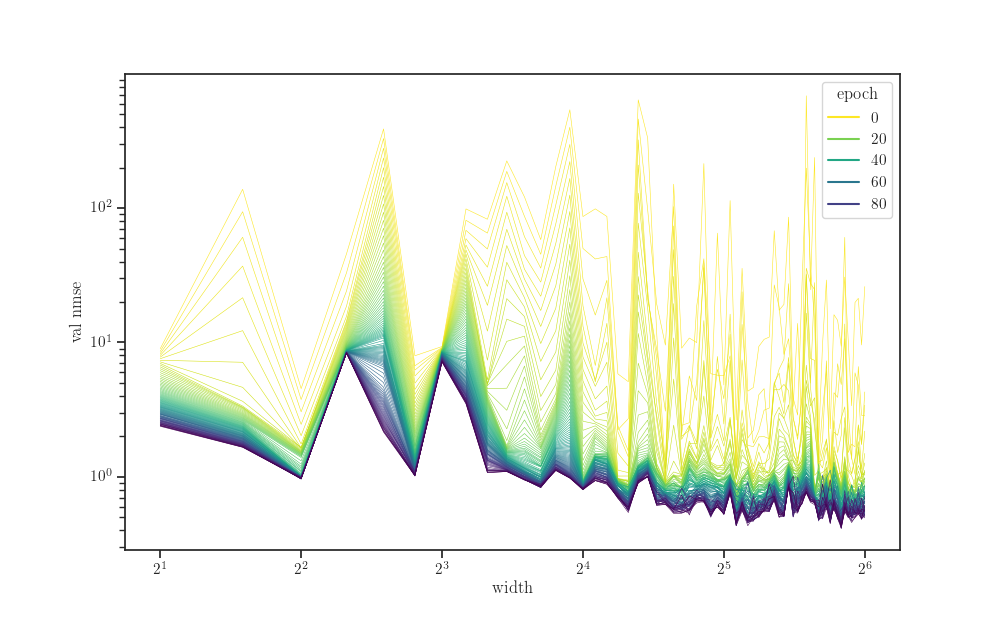
fig, ax = plt.subplots()
sns.lineplot(x="width", y="val nmse", hue="epoch",
units="seed", estimator=None,
palette="viridis_r", linewidth=0.4,
data=data, ax=ax)
ax.set_xscale("log", base=2)
ax.set_yscale("log")
plt.show()
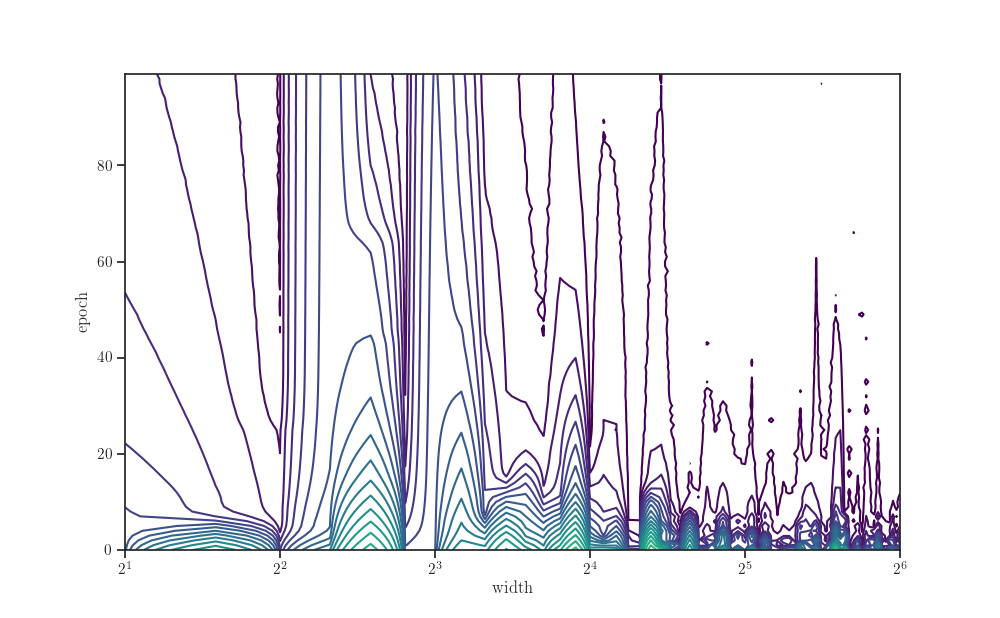
new_data = data.pivot(index="width", columns="epoch", values="val nmse")
Z = new_data.to_numpy()
fig, ax = plt.subplots()
ax.contour(*np.broadcast_arrays(width_grid.reshape(-1, 1), t_grid), Z,
levels=np.logspace(0, 4, 25), norm=LogNorm(), cmap="viridis")
ax.set_xscale("log", base=2)
ax.set_xlabel(r"width")
ax.set_ylabel(r"epoch")
plt.show()
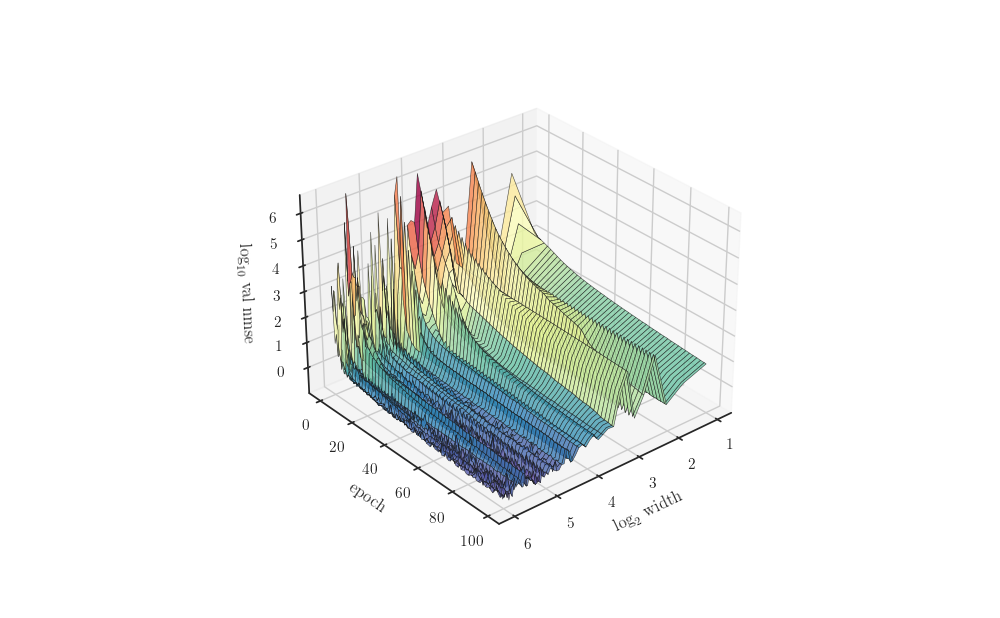
fig, ax = plt.subplots(subplot_kw=dict(projection="3d", azim=50))
ax.plot_surface(np.log2(width_grid).reshape(-1, 1), t_grid, np.log(Z), alpha=0.8,
edgecolor='k', linewidth=0.4, cmap="Spectral_r")
ax.set_xlabel(r"$\log_2$ width")
ax.set_ylabel("epoch")
ax.set_zlabel(r"$\log_{10}$ val nmse")
plt.show()
new_data.rename(lambda s: s + 1, axis="columns", inplace=True)
columns = list(np.minimum(3**np.arange(6), num_epochs))
g = sns.PairGrid(new_data[columns], corner=True)
g = g.map_lower(plt.plot)
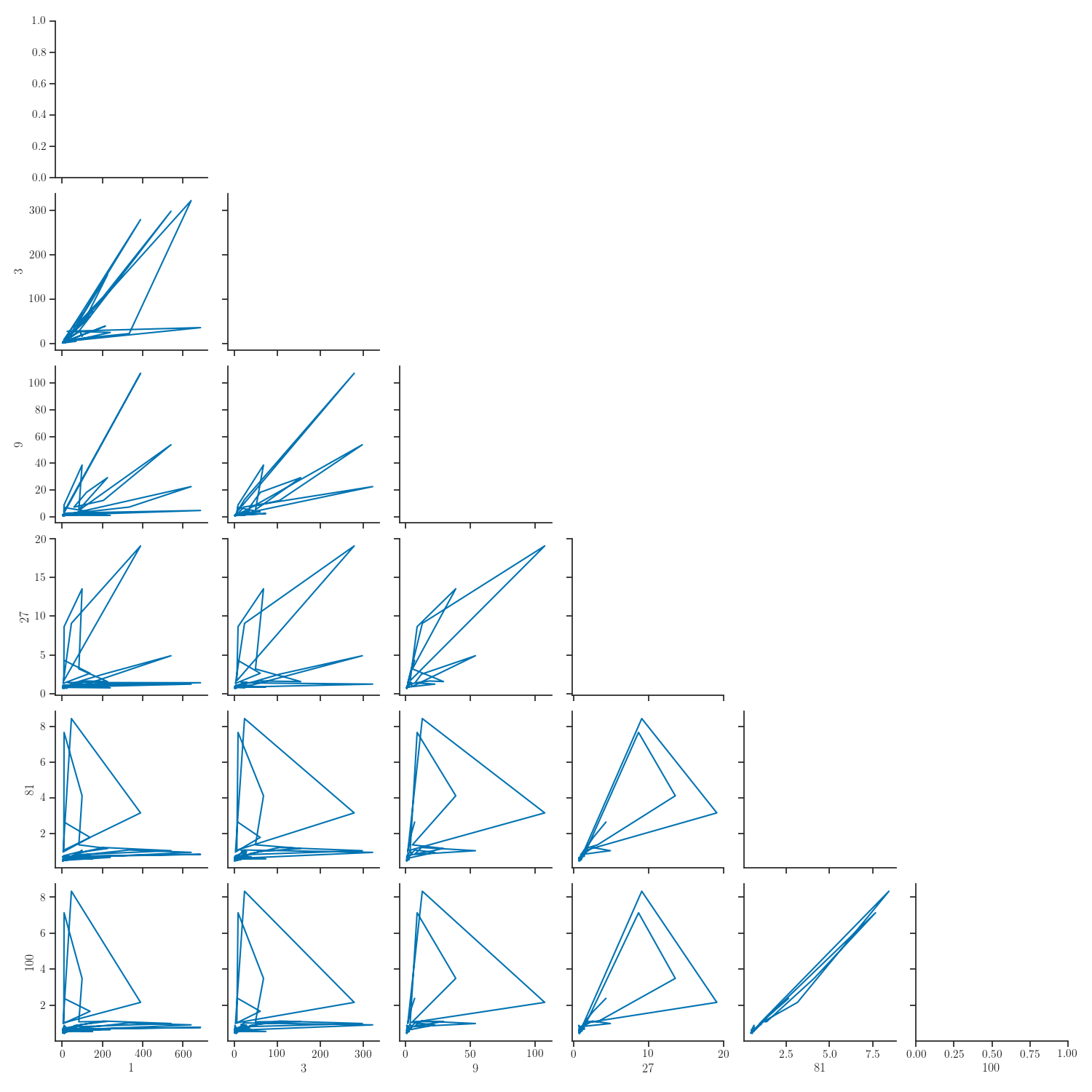
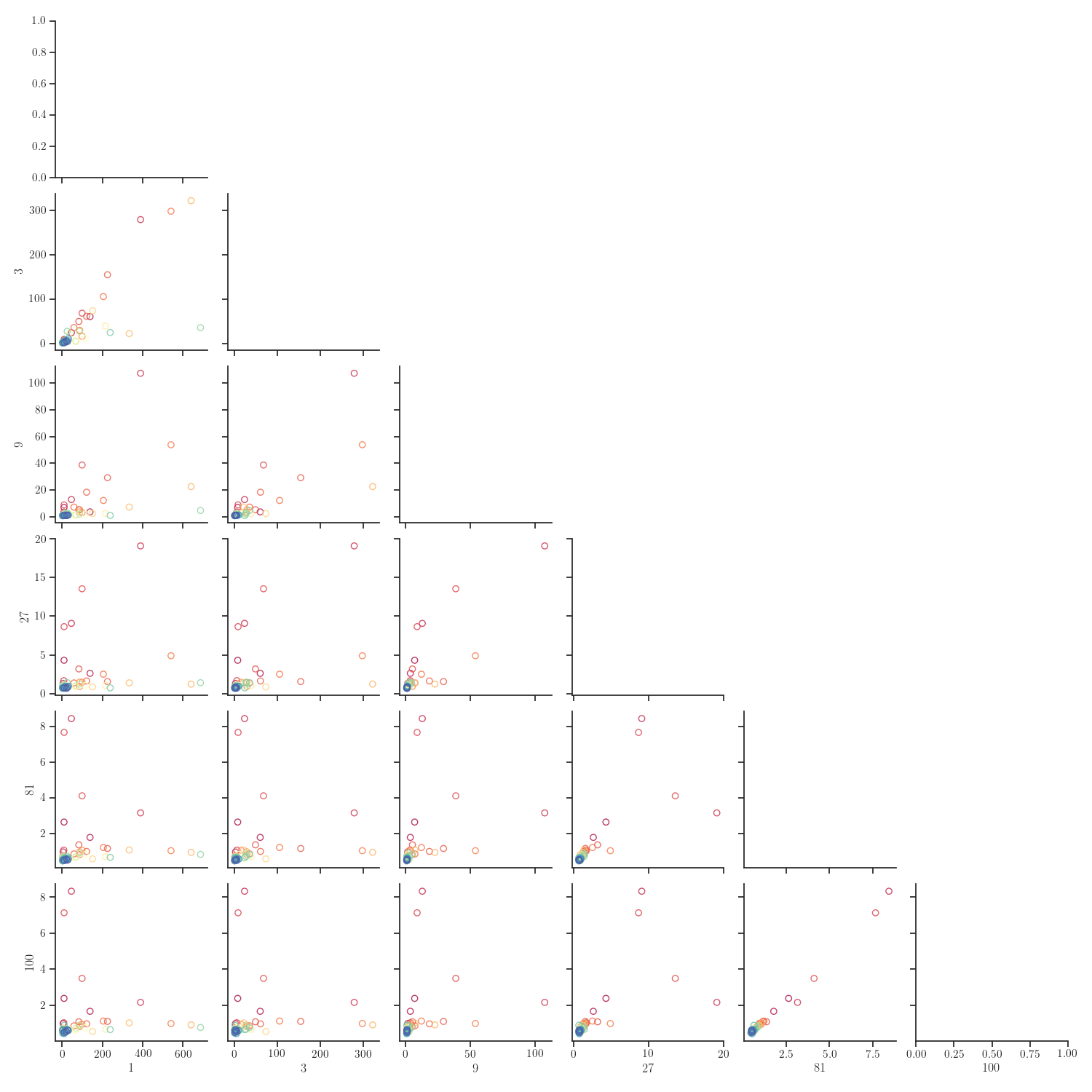
g = sns.PairGrid(new_data[columns].reset_index(),
hue="width", palette="Spectral", corner=True)
g = g.map_lower(plt.scatter, facecolor="none", alpha=0.8)
Total running time of the script: ( 3 minutes 6.121 seconds)