Note
Click here to download the full example code
Gaussian Process Posterior Predictive¶
Hello world
import numpy as np
import tensorflow_probability as tfp
import matplotlib.pyplot as plt
from etudes.datasets.synthetic import synthetic_sinusoidal, make_regression_dataset
from etudes.plotting import fill_between_stddev
# shortcuts
tfd = tfp.distributions
kernels = tfp.math.psd_kernels
# constants
num_train = 25 # nbr training points in synthetic dataset
num_features = 1 # dimensionality
num_index_points = 256 # nbr of index points
num_samples = 7
amplitude = 1.0
length_scale = 0.1
observation_noise_variance = 1e-1
jitter = 1e-6
kernel_cls = kernels.ExponentiatedQuadratic
seed = 42 # set random seed for reproducibility
random_state = np.random.RandomState(seed)
x_min, x_max = -1.0, 1.0
X_test = np.linspace(x_min, x_max, num_index_points).reshape(-1, num_features)
load_data = make_regression_dataset(synthetic_sinusoidal)
X_train, Y_train = load_data(num_train, num_features,
observation_noise_variance,
x_min=-0.5, x_max=0.5,
random_state=random_state)
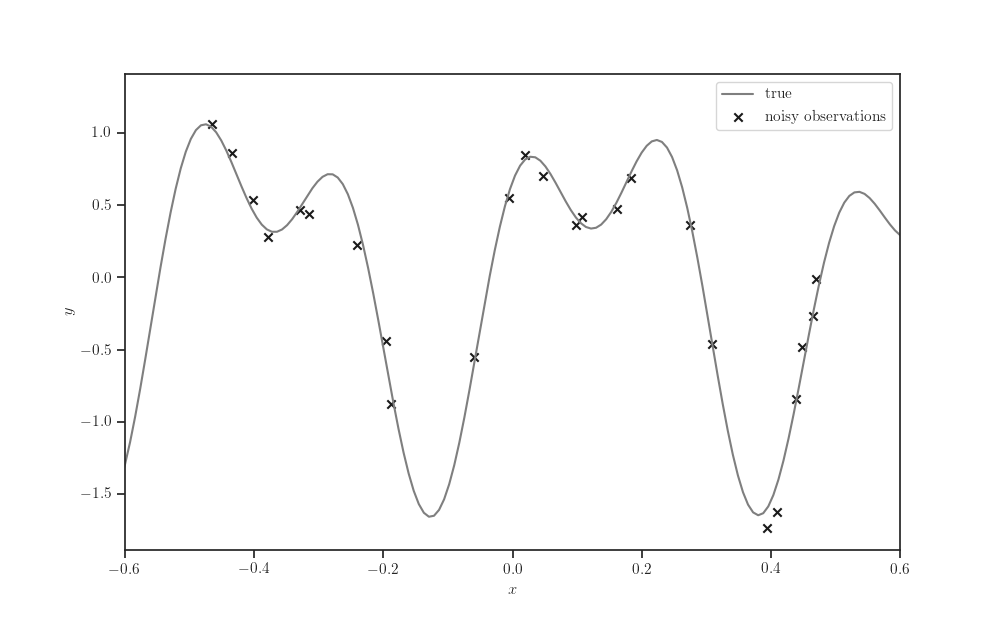
Synthetic dataset¶
fig, ax = plt.subplots()
ax.plot(X_test, synthetic_sinusoidal(X_test), label="true", color="tab:gray")
ax.scatter(X_train, Y_train, marker='x', color='k',
label="noisy observations")
ax.legend()
ax.set_xlim(-0.6, 0.6)
ax.set_xlabel('$x$')
ax.set_ylabel('$y$')
plt.show()
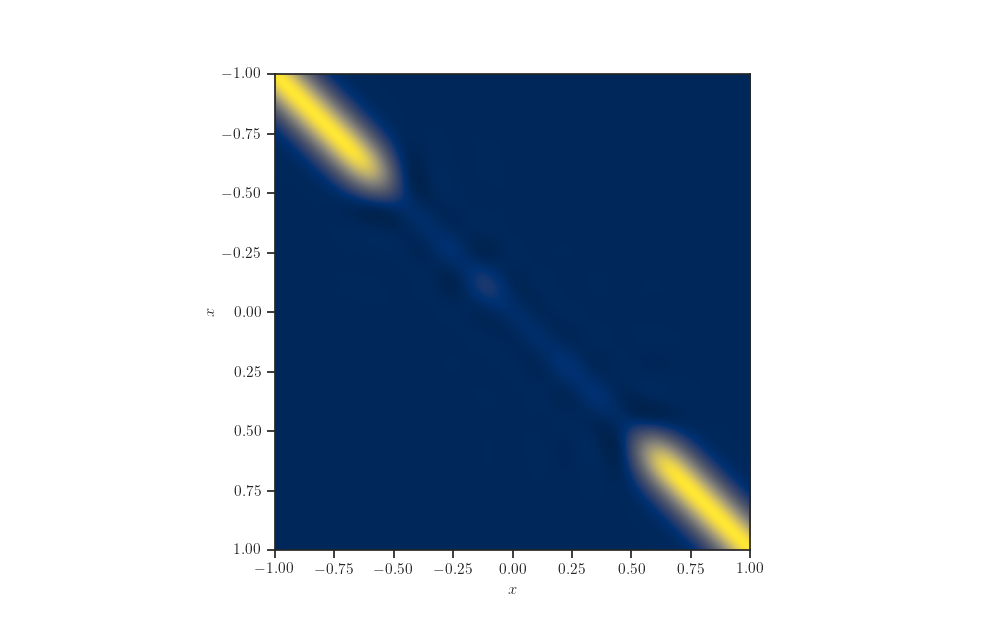
Schur Complement kernel matrix¶
kernel = kernel_cls(amplitude=np.float64(amplitude),
length_scale=np.float64(length_scale))
schur_complement_kernel = kernels.SchurComplement(
kernel, X_train, diag_shift=observation_noise_variance)
fig, ax = plt.subplots()
ax.imshow(schur_complement_kernel.matrix(X_test, X_test),
extent=[x_min, x_max, x_max, x_min], cmap="cividis")
ax.set_xlabel(r'$x$')
ax.set_ylabel(r'$x$')
plt.show()
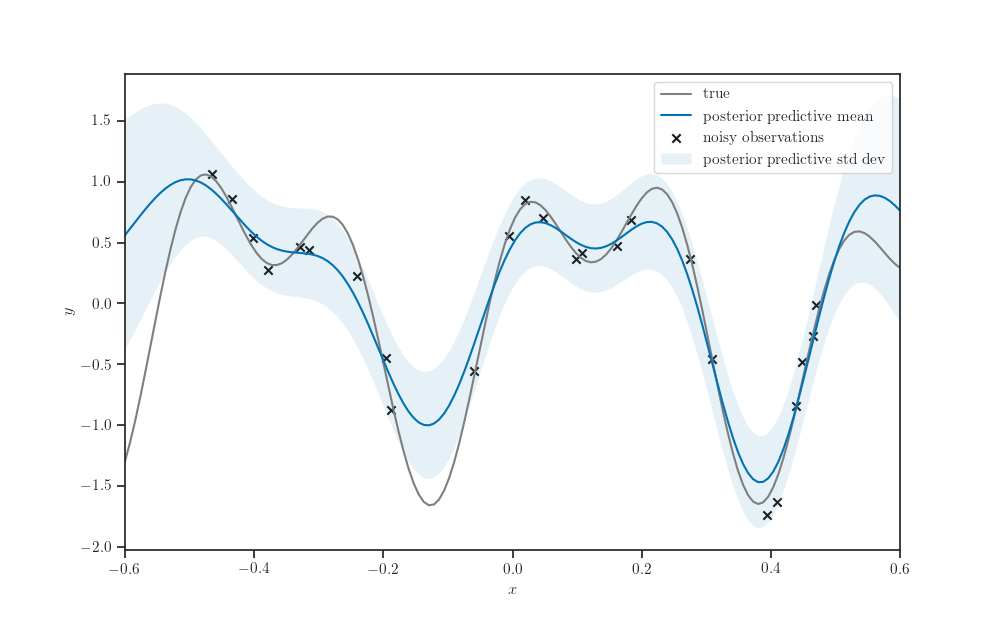
Posterior predictive distribution¶
gprm = tfd.GaussianProcessRegressionModel(
kernel=kernel, index_points=X_test,
observation_index_points=X_train,
observations=Y_train,
observation_noise_variance=observation_noise_variance,
jitter=jitter
)
fig, ax = plt.subplots()
ax.plot(X_test, synthetic_sinusoidal(X_test), label="true", color="tab:gray")
ax.scatter(X_train, Y_train, marker='x', color='k',
label="noisy observations")
ax.plot(X_test, gprm.mean(), label="posterior predictive mean")
fill_between_stddev(X_test.squeeze(),
gprm.mean().numpy().squeeze(),
gprm.stddev().numpy().squeeze(), alpha=0.1,
label="posterior predictive std dev", ax=ax)
ax.legend()
ax.set_xlim(-0.6, 0.6)
ax.set_xlabel('$x$')
ax.set_ylabel('$y$')
plt.show()
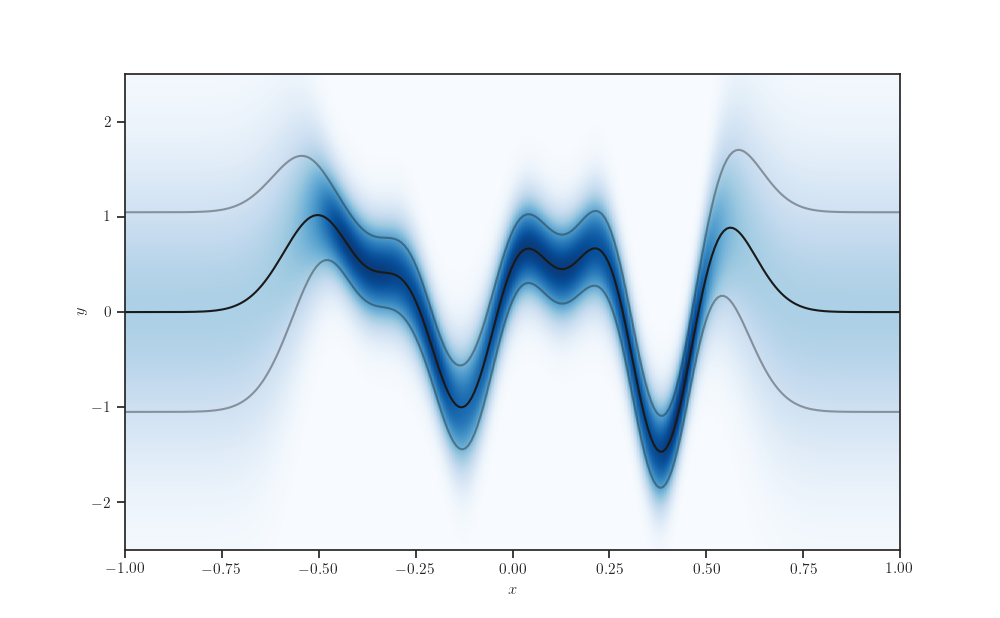
y_min, y_max = -2.5, 2.5
y_num_points = 512
gprm_marginals = tfd.Normal(loc=gprm.mean(), scale=gprm.stddev())
Y_q = np.linspace(y_min, y_max, y_num_points).reshape(-1, 1)
fig, ax = plt.subplots()
ax.imshow(gprm_marginals.prob(Y_q), origin="lower",
extent=[x_min, x_max, y_min, y_max],
aspect="auto", cmap="Blues")
ax.plot(X_test, gprm.mean(), color='k')
ax.plot(X_test, gprm.mean() - gprm.stddev(), color='k', alpha=0.4)
ax.plot(X_test, gprm.mean() + gprm.stddev(), color='k', alpha=0.4)
ax.set_xlabel('$x$')
ax.set_ylabel('$y$')
plt.show()
Total running time of the script: ( 0 minutes 5.740 seconds)